Survival rate of prostate cancer in a Sudameric Ocncologyc Instutute
Prostate cancer is the most common malignant tumor in men and the vast majority of cases occur after the age of 65 years and rarely in those younger than 45 years.
The survival rate is the percentage of people who survive after being diagnosed with a disease within a certain period of time (5 to 10 years). In general, prostate cancer survival rates are very good when the disease is diagnosed in early stages, reaching 100%, 98% and 93% at 5, 10 and 15 years, respectively. This rate drops to 30% at 5 years if the diagnosis is made in advanced stages. Therefore, the survival rate is directly related to the clinical stage of the disease.
The data for this small review were obtained from the medical records of 1639 patients treated in the period 2000-2010 in an Oncological Institution in South America specialized in cancer treatment. Therefore, patient data are confidential and have been anonymized.
The aim of this small review is to know what is the survival rate in this oncological institute for patients diagnosed in the period indicated above.
Exploratory Data Analysis
from dateutil.relativedelta import relativedelta
import matplotlib.pyplot as plt
from datetime import datetime
import seaborn as sns
import pandas as pd
import numpy as np
import matplotlib
import math
import re
1.- Read data
df_ca=pd.read_excel("patient_survival_ca_prostate_00-10.xlsx",
index_col=None,
na_values= np.nan)
To anonymize the database, the column containing the medical record number of all patients is removed. A new dataFarme is also created to assign each medical record to a number and, subsequently, to be able to trace each one.
df_final=df_ca.sort_values(["F.Diagnóstico"],ascending=True).reset_index()
df_HCL=df_final[["index","Num.HCL"]]
df_final.drop(["Num.HCL","Num_HCL"], axis=1,inplace=True)
2.- Data exploration
pd.set_option('display.max_columns', 15)
pd.set_option("max_colwidth", 18)
pd.set_option('display.max_rows', 22)
df_final.head()
| index | Edad Diag. | Cod Diag | Diagnóstico | Cod Sitio | Sitio | SEER Estadio | ... | Fec.Reg.Seguim. | Estado Vital | Clase Caso | Clase Caso.1 | F. Pase Control | E.Tratam. | Estado Tratamiento | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 59 | 81403 | Adenocarcinoma... | C619 | Glandula prost... | 9 | ... | 2017/01/27 | Vivo | 42 | Dx fuera y sol... | 2000/12/15 | 0 | Caso completo |
| 1 | 1 | 61 | 81403 | Adenocarcinoma... | C619 | Glandula prost... | 7 | ... | 2002/06/11 | Muerto | 32 | Dx y TODO el T... | 2001/04/20 | 0 | Caso completo |
| 2 | 2 | 74 | 81403 | Adenocarcinoma... | C619 | Glandula prost... | 7 | ... | 2012/03/07 | Muerto | 14 | Dx y TODO el T... | 2001/04/09 | 0 | Caso completo |
| 3 | 3 | 59 | 81403 | Adenocarcinoma... | C619 | Glandula prost... | 3 | ... | 2013/08/15 | Vivo | 14 | Dx y TODO el T... | 2000/05/08 | 0 | Caso completo |
| 4 | 4 | 62 | 81403 | Adenocarcinoma... | C619 | Glandula prost... | 2 | ... | 2013/06/04 | Muerto | 14 | Dx y TODO el T... | 2000/03/09 | 0 | Caso completo |
5 rows × 52 columns
df_final.tail()
| index | Edad Diag. | Cod Diag | Diagnóstico | Cod Sitio | Sitio | SEER Estadio | ... | Fec.Reg.Seguim. | Estado Vital | Clase Caso | Clase Caso.1 | F. Pase Control | E.Tratam. | Estado Tratamiento | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1634 | 1634 | 80 | 81403 | Adenocarcinoma... | C619 | Glandula prost... | 9 | ... | 2018/05/21 | Vivo | 22 | Dx fuera y TOD... | 2011/04/06 | 0 | Caso completo |
| 1635 | 1635 | 71 | 81403 | Adenocarcinoma... | C619 | Glandula prost... | 1 | ... | 2018/05/28 | Vivo | 22 | Dx fuera y TOD... | 2011/03/23 | 0 | Caso completo |
| 1636 | 1636 | 70 | 81403 | Adenocarcinoma... | C619 | Glandula prost... | 7 | ... | 2012/07/19 | Muerto | 14 | Dx y TODO el T... | 2011/03/01 | 0 | Caso completo |
| 1637 | 1637 | 61 | 81403 | Adenocarcinoma... | C619 | Glandula prost... | 9 | ... | 2018/05/16 | Vivo | 32 | Dx y TODO el T... | 2010/12/23 | 0 | Caso completo |
| 1638 | 1638 | 81 | 81403 | Adenocarcinoma... | C619 | Glandula prost... | 7 | ... | 2018/05/24 | Muerto | 22 | Dx fuera y TOD... | 2011/03/04 | 0 | Caso completo |
5 rows × 52 columns
df_final.describe()
| index | Edad Diag. | Cod Diag | SEER Estadio | TIPO CX Sitio Primario | Ciclos o Gray Recib.1 | Tipo Rec.1 | Tipo Rec.2 | Clase Caso | E.Tratam. | |
|---|---|---|---|---|---|---|---|---|---|---|
| count | 1639.000000 | 1639.000000 | 1639.000000 | 1639.000000 | 267.000000 | 326.000000 | 1639.000000 | 1639.000000 | 1639.000000 | 1639.000000 |
| mean | 819.000000 | 69.472849 | 81474.109823 | 5.082367 | 48.696629 | 68.644172 | 62.370958 | 838.461257 | 22.038438 | 0.768761 |
| std | 473.282861 | 8.768998 | 674.772300 | 3.323969 | 10.529305 | 8.998257 | 81.952525 | 199.876402 | 9.756766 | 19.205771 |
| min | 0.000000 | 35.000000 | 80003.000000 | 0.000000 | 5.000000 | 12.000000 | 0.000000 | 10.000000 | 0.000000 | 0.000000 |
| 25% | 409.500000 | 63.000000 | 81403.000000 | 2.000000 | 50.000000 | 70.000000 | 10.000000 | 888.000000 | 14.000000 | 0.000000 |
| 50% | 819.000000 | 70.000000 | 81403.000000 | 7.000000 | 50.000000 | 70.000000 | 70.000000 | 888.000000 | 22.000000 | 0.000000 |
| 75% | 1228.500000 | 76.000000 | 81403.000000 | 9.000000 | 50.000000 | 70.000000 | 99.000000 | 888.000000 | 22.000000 | 0.000000 |
| max | 1638.000000 | 96.000000 | 96803.000000 | 9.000000 | 99.000000 | 120.000000 | 888.000000 | 888.000000 | 99.000000 | 777.000000 |
df_final.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1639 entries, 0 to 1638
Data columns (total 52 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 index 1639 non-null int64
1 Edad Diag. 1639 non-null int64
2 Cod Diag 1639 non-null int64
3 Diagnóstico 1639 non-null object
4 Cod Sitio 1639 non-null object
5 Sitio 1639 non-null object
6 SEER Estadio 1639 non-null int64
7 SEER Estadio.1 1639 non-null object
8 TNM Estadio RH 1639 non-null object
9 Otra Extensión 1639 non-null object
10 F.Diagnóstico 1639 non-null object
11 Tto.afuera 282 non-null object
12 F.Tto.afuera 342 non-null object
13 Ttto no curativos 419 non-null object
14 F.Ttto no curat 421 non-null object
15 Fecha 1erTto. - Razón 806 non-null object
16 F.Aband.Tto 217 non-null object
17 Fecha CIRUGIA 277 non-null object
18 Razón PARA NO CX 277 non-null object
19 TIPO CX Sitio Primario 267 non-null float64
20 fecha SIN TTO CLINICO 993 non-null object
21 razon PARA SIN TTO CLINICO 993 non-null object
22 Tipo RADIOTERAPIA 328 non-null object
23 Fecha RT 340 non-null object
24 Razón PARA NO RT 340 non-null object
25 Ciclos o Gray Recib.1 326 non-null float64
26 Fecha HORMONOTERAPIA 129 non-null object
27 Razón PARA NO HT 129 non-null object
28 Fecha ORQUIECTOMIA 235 non-null object
29 Razón PARA NO ORQUIECTOMIA 235 non-null object
30 Tipo OTROS TTOS 7 non-null object
31 Fecha OTROS TTOS 7 non-null object
32 Razón PARA NO OTROS TTOS 7 non-null object
33 Metast.A Rec. 1639 non-null object
34 Metast.B Rec. 1639 non-null object
35 Metast.C Rec. 1639 non-null object
36 Tipo Rec.1 1639 non-null int64
37 Tipo Rec.2 1639 non-null int64
38 F.Recurrencia 1182 non-null object
39 F.Tto Recurrencia 418 non-null object
40 Tipos Tto Recurrencia 976 non-null object
41 Fecha Defun. 871 non-null object
42 C.Defun. 1639 non-null object
43 Causa Defunción 870 non-null object
44 Fec.Ult.Contacto 1639 non-null object
45 Fec.Reg.Seguim. 1639 non-null object
46 Estado Vital 1639 non-null object
47 Clase Caso 1639 non-null int64
48 Clase Caso.1 1639 non-null object
49 F. Pase Control 1624 non-null object
50 E.Tratam. 1639 non-null int64
51 Estado Tratamiento 1639 non-null object
dtypes: float64(2), int64(8), object(42)
memory usage: 666.0+ KB
df_final.isnull().sum()[df_final.isnull().sum() !=0]
Tto.afuera 1357
F.Tto.afuera 1297
Ttto no curativos 1220
F.Ttto no curat 1218
Fecha 1erTto. - Razón 833
...
F.Tto Recurrencia 1221
Tipos Tto Recurrencia 663
Fecha Defun. 768
Causa Defunción 769
F. Pase Control 15
Length: 28, dtype: int64
df_final.groupby(['Diagnóstico']).count()\
.assign(Count=lambda dataset:dataset['Edad Diag.'],
Percentage=lambda dataset:dataset['Edad Diag.']*100/dataset['Edad Diag.'].sum(),
)[["Count","Percentage"]].sort_values("Count",ascending=False)
| Count | Percentage | |
|---|---|---|
| Diagnóstico | ||
| Adenocarcinoma SAI | 1591 | 97.071385 |
| Carcinoma de cel.acinosas | 25 | 1.525320 |
| Neo. maligna | 7 | 0.427090 |
| Adenocar.tubular | 2 | 0.122026 |
| Ca.de cel.transicionales SAI | 2 | 0.122026 |
| Carcinoma SAI | 2 | 0.122026 |
| Carcinoma indiferenciado SAI | 2 | 0.122026 |
| Carcinoma neuroendocrino SAI | 2 | 0.122026 |
| Adenocar. mucinoso | 1 | 0.061013 |
| Adenocarcinoma .de cel. claras SAI | 1 | 0.061013 |
| Carcinoma de cel.pequenas SAI Neuroendocrino | 1 | 0.061013 |
| Carcinoma in situ SAI | 1 | 0.061013 |
| Leiomiosarcoma SAI | 1 | 0.061013 |
| Linfoma maglino cels B grandes difuso SAI | 1 | 0.061013 |
3.- Data cleaning
Because a review of the survival rate is to be performed, columns that are not necessary are eliminated.
delete_keys=['Cod Diag','Cod Sitio','Sitio','F.Ttto no curat',
'Razón PARA NO CX', 'TIPO CX Sitio Primario',
'fecha SIN TTO CLINICO','Razón PARA NO RT',
'Razón PARA NO HT','Tipo OTROS TTOS','Razón PARA NO ORQUIECTOMIA',
'Fecha OTROS TTOS', 'Razón PARA NO OTROS TTOS',
'Metast.B Rec.', 'Metast.C Rec.','C.Defun.',
'Clase Caso','Clase Caso.1','Num_HCL','Otra Extensión']
target_keys=[item for item in df_final.keys() if item not in delete_keys]
df_final=df_final[target_keys]
df_final.head()
| index | Edad Diag. | Diagnóstico | SEER Estadio | SEER Estadio.1 | TNM Estadio RH | F.Diagnóstico | ... | Causa Defunción | Fec.Ult.Contacto | Fec.Reg.Seguim. | Estado Vital | F. Pase Control | E.Tratam. | Estado Tratamiento | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 59 | Adenocarcinoma... | 9 | No estadificad... | T777; N777; M7... | 2000/01/02 | ... | NaN | 2014/06/16 | 2017/01/27 | Vivo | 2000/12/15 | 0 | Caso completo |
| 1 | 1 | 61 | Adenocarcinoma... | 7 | Metastasis dis... | T777; N777; M7... | 2000/01/04 | ... | Tumor maligno ... | 2001/04/20 | 2002/06/11 | Muerto | 2001/04/20 | 0 | Caso completo |
| 2 | 2 | 74 | Adenocarcinoma... | 7 | Metastasis dis... | TX; NX; MX; EIV | 2000/01/05 | ... | Enfermedades d... | 2010/07/15 | 2012/03/07 | Muerto | 2001/04/09 | 0 | Caso completo |
| 3 | 3 | 59 | Adenocarcinoma... | 3 | Regional A Los... | T777; N777; M7... | 2000/01/05 | ... | NaN | 2011/08/09 | 2013/08/15 | Vivo | 2000/05/08 | 0 | Caso completo |
| 4 | 4 | 62 | Adenocarcinoma... | 2 | Regional Por E... | TX; NX; MX; EIV | 2000/01/13 | ... | Sintomas/ sign... | 2002/12/12 | 2013/06/04 | Muerto | 2000/03/09 | 0 | Caso completo |
5 rows × 33 columns
Now, the names of variables (columns) whose format makes correct data manipulation impossible are changed.
df_final=df_final.rename(columns={'Edad Diag.':'Edad_diag',
'SEER Estadio':'Cod_SEER_Estadio',
'SEER Estadio.1':'SEER_Estadio',
'Diagnóstico': 'Diagnostico',
'TNM Estadio RH':'TNM_Estadio_RH',
'F.Diagnóstico':'Fecha_Diag',
'Tto.afuera':'Tto_afuera',
'F.Tto.afuera':'Fecha_Tto_afuera',
'Ttto no curativos':'Ttto_paliativo',
'Fecha 1erTto. - Razón':'Tto_1',
'F.Aband.Tto':'Fecha_Aband_Tto',
'Fecha CIRUGIA':'Fecha_CX',
'razon PARA SIN TTO CLINICO':'Muere_antes_Tto',
'Tipo RADIOTERAPIA':'Radioterapia',
'Fecha RT':'Fecha_RT',
'Ciclos o Gray Recib.1':'Dosis_Recib',
'Fecha HORMONOTERAPIA':'Fecha_HT',
'Fecha ORQUIECTOMIA':'Fecha_orquiectomia',
'Metast.A Rec.':'Metastasis',
'Tipo Rec.1':'Tipo_Rec_1',
'Tipo Rec.2':'Tipo_Rec_2',
'F.Recurrencia':'Fecha_Rec',
'F.Tto Recurrencia':'Fecha_Tto_Rec',
'Tipos Tto Recurrencia':'Tipos_Tto_Rec',
'Fecha Defun.':'Fecha_Defun',
'Causa Defunción':'Causa_Defuncion',
'Fec.Ult.Contacto':'Fecha_Ult_Contacto',
'Fec.Reg.Seguim.':'Fecha_Reg_Seguim',
'Estado Vital':'Estado_Vital',
'F. Pase Control':'Fecha_Pase_Control',
'E.Tratam.':'Cod_Estado_Tratam',
'Estado Tratamiento':'Estado_Tratamiento'})
In order to be able to manipulate the data through dataframe.assign(), the “na” values are replaced with dataframe.fillna() by a blank space (“”).
df_final=df_final.fillna("")
Continuing with the cleaning we notice that there are 4 variables contained in one (“TNM_Estadio_RH”), which are in string format, so this string is divided with str.split() considering “;” as separator.The value of each is stored in a new variable. Finally, the column “TNM_State_RH” is deleted.
dp=df_final.assign(T=lambda dataset:dataset["TNM_Estadio_RH"]\
.apply(lambda row:row.split(sep=';')[0]\
.split(sep='T')[1]),
N=lambda dataset:dataset["TNM_Estadio_RH"]\
.apply(lambda row:row.split(sep=';')[1]\
.split(sep='N')[1]),
M=lambda dataset:dataset["TNM_Estadio_RH"]\
.apply(lambda row:row.split(sep=';')[2]\
.split(sep='M')[1]),
E=lambda dataset:dataset["TNM_Estadio_RH"]\
.apply(lambda row:row.split(sep=';')[3]\
.split(sep='E')[1])
).drop(["TNM_Estadio_RH"], axis=1)
Now, there is also the variables “Tto_afuera” and “Tipos_Tto_Rec” which has several variables contained in it. TThese variables will be created as dummy variables to represent their presence or absence.
dp=dp.assign(CX_fuera=lambda dataset:dataset["Tto_afuera"]\
.apply(lambda row:1 if re.search('CX',row)\
else 0),
HT_fuera=lambda dataset:dataset["Tto_afuera"]\
.apply(lambda row:1 if re.search('HT',row)\
else 0),
QT_fuera=lambda dataset:dataset["Tto_afuera"]\
.apply(lambda row:1 if re.search('QT',row)\
else 0),
OT_fuera=lambda dataset:dataset["Tto_afuera"]\
.apply(lambda row:1 if re.search('OT',row)\
else 0),
CX_Rec=lambda dataset:dataset["Tipos_Tto_Rec"]\
.apply(lambda row:1 if re.search('CX',row)\
else 0),
HT_Rec=lambda dataset:dataset["Tipos_Tto_Rec"]\
.apply(lambda row:1 if re.search('HT',row)\
else 0),
QT_Rec=lambda dataset:dataset["Tipos_Tto_Rec"]\
.apply(lambda row:1 if re.search('QT',row)\
else 0),
RT_Rec=lambda dataset:dataset["Tipos_Tto_Rec"]\
.apply(lambda row:1 if re.search('RT',row)\
else 0)
).drop(["Tipos_Tto_Rec"], axis=1)
The value of the variable “Fecha_Tto_1” contains values of two different variables separated by a hyphen. This variable expresses the treatment start date followed by the type of treatment.
dp=dp.assign(Fecha_Tto_1=lambda dataset:dataset["Tto_1"]\
.apply(lambda row:row.split(sep='-')[0]),
Tto_1=lambda dataset:dataset["Tto_1"]\
.apply(lambda row: re.search(r'[a-zA-Z0]+$',row)[0]
if re.search(r'[a-zA-Z0]+$',row)
else row))
Since there is only the variable with the date of the patients who underwent a certain procedure/treatment, a new variable is created containing information on the presence or absence of the procedure/treatment.Invalid values must be considered:
- ” “ : Blank space (absence of information)
- 777 : No information available
- 888 : Not applicable
dp=dp.assign(Aband_Tto=lambda dataset:dataset["Fecha_Aband_Tto"]\
.apply(lambda row: 1 if row!=""and row!="888" and row!="777"\
else 0),
Hormonoterapia=lambda dataset:dataset["Fecha_HT"]\
.apply(lambda row: 1 if row!=""and row!="888" and row!="777"\
else 0),
Orquiectomia=lambda dataset:dataset["Fecha_orquiectomia"]\
.apply(lambda row: 1 if row!=""and row!="888" and row!="777"\
else 0),
Recurrecia=lambda dataset:dataset["Fecha_Rec"]\
.apply(lambda row: 1 if row!=""and row!="888" and row!="777"\
else 0)
)
In the variable “Muere_antes_Tto” there are several codes that represent different events of which we only need to know if the patient died before the treatment was started, so we replace them.
dp=dp.assign(Muere_antes_Tto=lambda dataset:dataset["Muere_antes_Tto"]\
.apply(lambda row:1 if row=='ST02'\
else 0))
A binary value representing the presence or absence of treatment is used to determine whether the patient received a certain treatment. This type of value is also used to determine whether or not the cancer has spread in the patient. Invalid values must be considered:
- ” “ : Blank space (absence of information)
- 777 : No information available
- 888 : Not applicable
dp=dp.assign(Ttto_paliativo=lambda dataset:dataset["Ttto_paliativo"]\
.apply(lambda row:1 if row!=""and row!="888" and row!="777"\
else 0),
Radioterapia=lambda dataset:dataset["Radioterapia"]\
.apply(lambda row:1 if re.search(r'^RT',row)\
else 0),
Metastasis=lambda dataset:dataset["Metastasis"]\
.apply(lambda row:1 if row!="" and row!="888" and row!="777"\
else 0)
)
In order to respect the SEER classification that classifies the stage of the patients, we replace only with the categories obtained from the official website in which there is a table of Summary Stage SS2018 of prostate cancer with their respective weighting.
dp=dp.assign(SEER_Estadio=lambda dataset:dataset["SEER_Estadio"].replace(
{
"No estadificado/ desconoce/ no especificado":"Desconocido",
"Metastasis distante / enferm. sistematica":"Distante",
"Regional Por Extension Directa":"Regional sólo por extensión directa",
"Regional A Los Ganglios Linfaticos":"Regional solo por Ganglios Linfaticos",
'Regional NEO':'Regional (2 y 3 )'
}
))
Also, in order to respect the SEER stadium coding, the values that do not belong are replaced by their corresponding value obtained from Summary Stage 2018: Prostate
dp["Cod_SEER_Estadio"].unique()
array([9, 7, 3, 2, 1, 0, 4, 5])
dp[~dp["Cod_SEER_Estadio"].isin([9,7,4,3,2,1,0])]
| index | Edad_diag | Diagnostico | Cod_SEER_Estadio | SEER_Estadio | Fecha_Diag | Tto_afuera | ... | QT_Rec | RT_Rec | Fecha_Tto_1 | Aband_Tto | Hormonoterapia | Orquiectomia | Recurrecia | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1501 | 1501 | 52 | Adenocarcinoma... | 5 | Regional (2 y 3 ) | 2010/04/20 | CX | ... | 0 | 0 | 2010/09/13 | 0 | 0 | 0 | 1 |
1 rows × 48 columns
All values different from 9,7,4,3,2,1 and 0, must be replaced with their corresponding value obtained in the page mentioned above.
dp=dp.assign(Cod_SEER_Estadio=lambda dataset:dataset["Cod_SEER_Estadio"].replace(
{
5:4
})
)
The [“Causa_Defuncion”] variable must keep the categories related to causes that indicate the influence of cancer on it, so the categories that have no relevance are replaced by “Otros”.
dp2=dp.assign(Causa_Defuncion=lambda dataset:dataset["Causa_Defuncion"].replace(
{
"Sintomas/ signos y hallazgos ":"Otros",
"Tumor maligno de Organos digestivos":"Otros",
"Enfermedades del sistema circulatorio":"Otros",
"Enfermedades del aparato digestivo":"Otros",
"Enfermedades endocrinas/ ":"Otros",
"Tumores malignos tejido linfatico/ de ":"Tumores malignos tejido linfatico",
"Tumor maligno de Piel" :"Otros",
"Tumor maligno de Labio/ cavidad ":"Otros",
"sin dato":np.nan,
"Tumores malignos de sitios mal ":"Otros",
"Enfermedades de la sangre y organos " :"Otros",
"Tumores malignos (primarios) de " :"Otros",
"Causas extremas de morbilidad y de " :"Otros",
"Tumor maligno de Ojo/ encefalo y " :"Otros",
"Tumor maligno de Tejidos " :"Otros"
}
))
The categories in the column [“Causa_Defuncion”] are checked using dataFrame.Series.value_counts()
dp2["Causa_Defuncion"].value_counts()
769
Tumor maligno de Organos genitales 582
Otros 262
Tumor maligno de Vias urinarias 9
Enfermedades del sistema 7
Tumores malignos tejido linfatico 5
Enfermedades del aparato 3
Name: Causa_Defuncion, dtype: int64
Now, we check the format of the variables using dataFrame.info()
dp.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1639 entries, 0 to 1638
Data columns (total 48 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 index 1639 non-null int64
1 Edad_diag 1639 non-null int64
2 Diagnostico 1639 non-null object
3 Cod_SEER_Estadio 1639 non-null int64
4 SEER_Estadio 1639 non-null object
5 Fecha_Diag 1639 non-null object
6 Tto_afuera 1639 non-null object
7 Fecha_Tto_afuera 1639 non-null object
8 Ttto_paliativo 1639 non-null int64
9 Tto_1 1639 non-null object
10 Fecha_Aband_Tto 1639 non-null object
11 Fecha_CX 1639 non-null object
12 Muere_antes_Tto 1639 non-null int64
13 Radioterapia 1639 non-null int64
14 Fecha_RT 1639 non-null object
15 Dosis_Recib 1639 non-null object
16 Fecha_HT 1639 non-null object
17 Fecha_orquiectomia 1639 non-null object
18 Metastasis 1639 non-null int64
19 Tipo_Rec_1 1639 non-null int64
20 Tipo_Rec_2 1639 non-null int64
21 Fecha_Rec 1639 non-null object
22 Fecha_Tto_Rec 1639 non-null object
23 Fecha_Defun 1639 non-null object
24 Causa_Defuncion 1639 non-null object
25 Fecha_Ult_Contacto 1639 non-null object
26 Fecha_Reg_Seguim 1639 non-null object
27 Estado_Vital 1639 non-null object
28 Fecha_Pase_Control 1639 non-null object
29 Cod_Estado_Tratam 1639 non-null int64
30 Estado_Tratamiento 1639 non-null object
31 T 1639 non-null object
32 N 1639 non-null object
33 M 1639 non-null object
34 E 1639 non-null object
35 CX_fuera 1639 non-null int64
36 HT_fuera 1639 non-null int64
37 QT_fuera 1639 non-null int64
38 OT_fuera 1639 non-null int64
39 CX_Rec 1639 non-null int64
40 HT_Rec 1639 non-null int64
41 QT_Rec 1639 non-null int64
42 RT_Rec 1639 non-null int64
43 Fecha_Tto_1 1639 non-null object
44 Aband_Tto 1639 non-null int64
45 Hormonoterapia 1639 non-null int64
46 Orquiectomia 1639 non-null int64
47 Recurrecia 1639 non-null int64
dtypes: int64(22), object(26)
memory usage: 614.8+ KB
Each variable representing a date must be changed to “datetime64[ns]” format.
dp2=dp2.assign(Fecha_Diag=lambda dataset:dataset["Fecha_Diag"].astype("datetime64[ns]"),
Fecha_Tto_afuera=lambda dataset:dataset["Fecha_Tto_afuera"]\
.astype("datetime64[ns]"),
Fecha_Tto_1=lambda dataset:dataset["Fecha_Tto_1"]\
.astype("datetime64[ns]"),
Fecha_Aband_Tto=lambda dataset:dataset["Fecha_Aband_Tto"]\
.astype("datetime64[ns]"),
Fecha_CX=lambda dataset:dataset["Fecha_CX"].astype("datetime64[ns]"),
Fecha_RT=lambda dataset:dataset["Fecha_RT"].astype("datetime64[ns]"),
Fecha_HT=lambda dataset:dataset["Fecha_HT"].astype("datetime64[ns]"),
Fecha_orquiectomia=lambda dataset:dataset["Fecha_orquiectomia"]\
.astype("datetime64[ns]"),
Fecha_Rec=lambda dataset:dataset["Fecha_Rec"].astype("datetime64[ns]"),
Fecha_Tto_Rec=lambda dataset:dataset["Fecha_Tto_Rec"]\
.astype("datetime64[ns]"),
Fecha_Defun=lambda dataset:dataset["Fecha_Defun"]\
.astype("datetime64[ns]"),
Fecha_Ult_Contacto=lambda dataset:dataset["Fecha_Ult_Contacto"]\
.astype("datetime64[ns]"),
Fecha_Reg_Seguim=lambda dataset:dataset["Fecha_Reg_Seguim"]\
.astype("datetime64[ns]"),
Fecha_Pase_Control=lambda dataset:dataset["Fecha_Pase_Control"]\
.astype("datetime64[ns]"),
)
For this type of review, only the stages are needed and not their subclassifications. A search is made for all the categories present in column “E”, which contains the stages of each patient.
dp2["E"].value_counts()
IV 628
777 350
II 268
III 202
99 100
I 84
IIIB 4
88 2
IC 1
Name: E, dtype: int64
Only stages I, II, III and IV are retained, and their subclassifications are attached to the main branch of stages.
dp2 = dp2.replace('IC', 'I')
dp2 = dp2.replace('IIIB', 'III')
Finally we replace the missing data with np.nan and rearrange the position of the columns.. Invalid values must be considered:
- ” “ : Blank space (absence of information)
- 777 : No information available
- 888 : Not applicable
- 99 : Not data
- 88 : Not data
dp2 = dp2.replace(['888','777','88','99',''], np.nan)
dp2 = dp2[[ "index","Edad_diag", "Diagnostico", "Cod_SEER_Estadio", "SEER_Estadio",
"Fecha_Diag", "T", "N", "M", "E", "CX_fuera", "HT_fuera",
"QT_fuera", "OT_fuera","Fecha_Tto_afuera", "Ttto_paliativo",
"Tto_1", "Muere_antes_Tto","Fecha_Tto_1","Aband_Tto",
"Fecha_Aband_Tto","Fecha_CX", "Hormonoterapia","Fecha_HT",
"Orquiectomia","Fecha_orquiectomia","Radioterapia","Dosis_Recib",
"Fecha_RT","Recurrecia","Tipo_Rec_1", "Tipo_Rec_2","Fecha_Rec",
"Metastasis","CX_Rec", "HT_Rec", "QT_Rec", "RT_Rec",
"Fecha_Tto_Rec","Fecha_Pase_Control","Fecha_Reg_Seguim",
"Fecha_Ult_Contacto","Cod_Estado_Tratam","Estado_Tratamiento",
"Estado_Vital","Fecha_Defun","Causa_Defuncion"]]
dp2.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1639 entries, 0 to 1638
Data columns (total 47 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 index 1639 non-null int64
1 Edad_diag 1639 non-null int64
2 Diagnostico 1639 non-null object
3 Cod_SEER_Estadio 1639 non-null int64
4 SEER_Estadio 1639 non-null object
5 Fecha_Diag 1639 non-null datetime64[ns]
6 T 1176 non-null object
7 N 1229 non-null object
8 M 1273 non-null object
9 E 1187 non-null object
10 CX_fuera 1639 non-null int64
11 HT_fuera 1639 non-null int64
12 QT_fuera 1639 non-null int64
13 OT_fuera 1639 non-null int64
14 Fecha_Tto_afuera 342 non-null datetime64[ns]
15 Ttto_paliativo 1639 non-null int64
16 Tto_1 806 non-null object
17 Muere_antes_Tto 1639 non-null int64
18 Fecha_Tto_1 806 non-null datetime64[ns]
19 Aband_Tto 1639 non-null int64
20 Fecha_Aband_Tto 217 non-null datetime64[ns]
21 Fecha_CX 277 non-null datetime64[ns]
22 Hormonoterapia 1639 non-null int64
23 Fecha_HT 129 non-null datetime64[ns]
24 Orquiectomia 1639 non-null int64
25 Fecha_orquiectomia 235 non-null datetime64[ns]
26 Radioterapia 1639 non-null int64
27 Dosis_Recib 326 non-null float64
28 Fecha_RT 340 non-null datetime64[ns]
29 Recurrecia 1639 non-null int64
30 Tipo_Rec_1 1639 non-null int64
31 Tipo_Rec_2 1639 non-null int64
32 Fecha_Rec 1182 non-null datetime64[ns]
33 Metastasis 1639 non-null int64
34 CX_Rec 1639 non-null int64
35 HT_Rec 1639 non-null int64
36 QT_Rec 1639 non-null int64
37 RT_Rec 1639 non-null int64
38 Fecha_Tto_Rec 418 non-null datetime64[ns]
39 Fecha_Pase_Control 1624 non-null datetime64[ns]
40 Fecha_Reg_Seguim 1639 non-null datetime64[ns]
41 Fecha_Ult_Contacto 1639 non-null datetime64[ns]
42 Cod_Estado_Tratam 1639 non-null int64
43 Estado_Tratamiento 1639 non-null object
44 Estado_Vital 1639 non-null object
45 Fecha_Defun 871 non-null datetime64[ns]
46 Causa_Defuncion 868 non-null object
dtypes: datetime64[ns](14), float64(1), int64(22), object(10)
memory usage: 601.9+ KB
Modification
New variables are created using the clean dataset in order to generate graphs to better visualize the contained data.
Two new variables are created in the dataset:
- tiempo_defuncion: years from diagnosis to death.
- tiempo_vivo: years from diagnosis to the time of database generation.
tiempo_final="2020-12-31"
tiempo_final=datetime.strptime(tiempo_final, '%Y-%m-%d')
dp2=dp2.assign(tiempo_defuncion=lambda dataset:round((dataset["Fecha_Defun"]-dataset["Fecha_Diag"]).dt.days / 365,1),
tiempo_vivo=lambda dataset:round((tiempo_final-dataset["Fecha_Diag"][dataset["Estado_Vital"]=="Vivo"]).dt.days / 365,1),
tiempo_recurr=lambda dataset:round((dataset["Fecha_Rec"]-dataset["Fecha_Diag"]).dt.days / 365,1))
A query is made to look for negative values in the new variable “tiempo_defuncion” since there should not be any value less than zero.
dp2[["Fecha_Diag","Fecha_Defun"]][dp2["tiempo_defuncion"]<0]
| Fecha_Diag | Fecha_Defun | |
|---|---|---|
| 493 | 2004-01-01 | 2001-02-11 |
| 1046 | 2007-10-09 | 2006-03-17 |
two negative values are found. Since there are very few data with respect to the whole dataset, they are eliminated.
dp2.drop(dp2[["Fecha_Diag","Fecha_Defun"]][dp2["tiempo_defuncion"]<0].index,inplace=True)
A query is made to look for negative values in the new variable “tiempo_vivo” since there should not be any value less than zero.
dp2[["Fecha_Diag","Fecha_Defun"]][dp2["tiempo_vivo"]<0]
| Fecha_Diag | Fecha_Defun |
|---|
Since no negative value is found, the datase is preserved. Finally, it can be said that the dataset is clean.
4.- Data visualization
sns.set_theme(style="darkgrid")
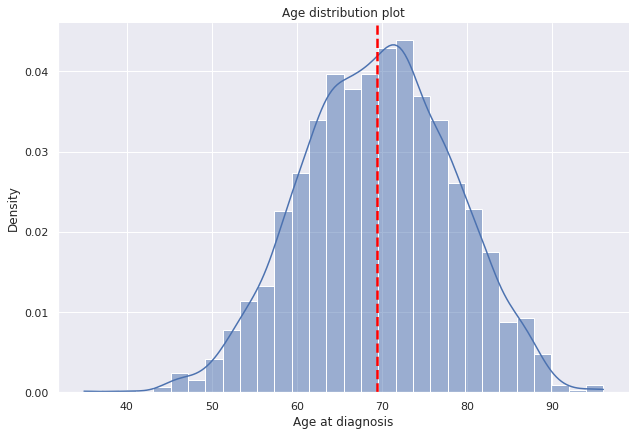
g=sns.displot(data=dp2,x="Edad_diag",
bins=30,kde=True,
kde_kws={'bw_adjust':0.9,'bw_method':'scott'},
stat='density', height=6,aspect=1.5).set(ylabel=None).set(title='Age distribution plot')
plt.axvline(dp2["Edad_diag"].mean(),c="red", ls='--',lw=2.5);
g.set(ylabel='Density', xlabel='Age at diagnosis');
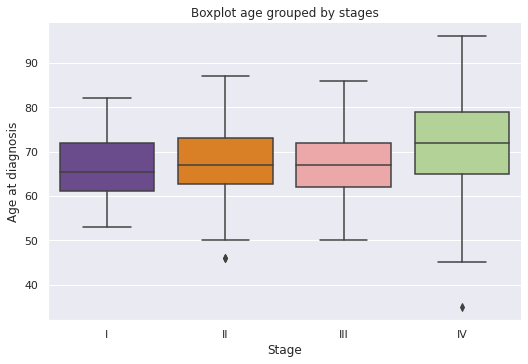
g = sns.catplot(x="E",
y="Edad_diag",
data=dp2.sort_values("E",ascending=True),
kind="box",
palette="Paired_r",
saturation=0.7,
height=5, aspect=1.5).set(title='Boxplot age grouped by stages')
g.set(xlabel='Stage', ylabel='Age at diagnosis');
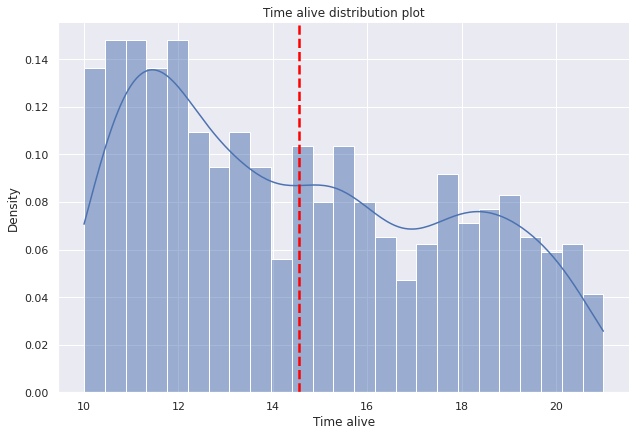
g=sns.displot(data=dp2,x="tiempo_vivo",
bins=25,kde=True,
kde_kws={'bw_adjust':0.9,'bw_method':'scott'},
stat='density', height=6,aspect=1.5).set(ylabel=None).set(title='Time alive distribution plot')
plt.axvline(dp2["tiempo_vivo"].mean(),c="red", ls='--',lw=2.5);
g.set(xlabel='Time alive', ylabel='Density');
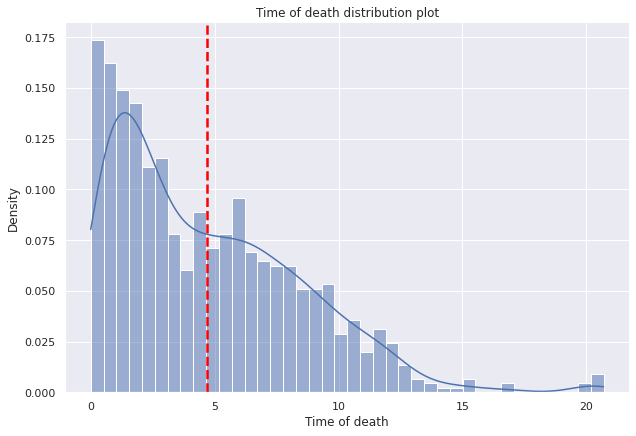
g=sns.displot(data=dp2,x="tiempo_defuncion",
bins=40,kde=True,
kde_kws={'bw_adjust':0.9,'bw_method':'scott'},
stat='density', height=6,aspect=1.5).set(ylabel=None).set(title='Time of death distribution plot')
plt.axvline(dp2["tiempo_defuncion"].mean(),c="red", ls='--',lw=2.5);
g.set(xlabel='Time of death', ylabel='Density');
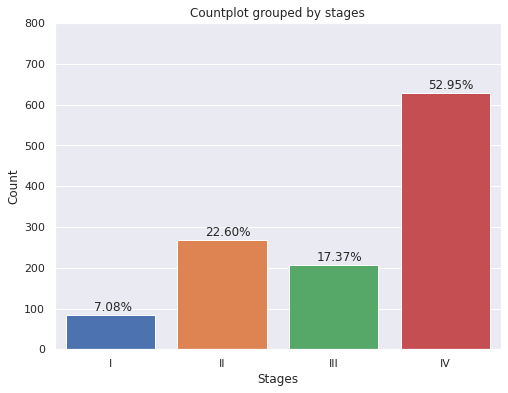
plt.figure(figsize = (8,6))
g=sns.countplot(x='E',
data=dp2.sort_values("E",ascending=True),
saturation=1)
g.set(ylim=(0, 800))
g.set(title='Countplot grouped by stages')
g.set(xlabel='Stages', ylabel='Count');
for p in g.patches:
g.annotate('{:.2f}%'.format(p.get_height()*100/dp2['E'].count()), (p.get_x()+0.25, p.get_height()+10))
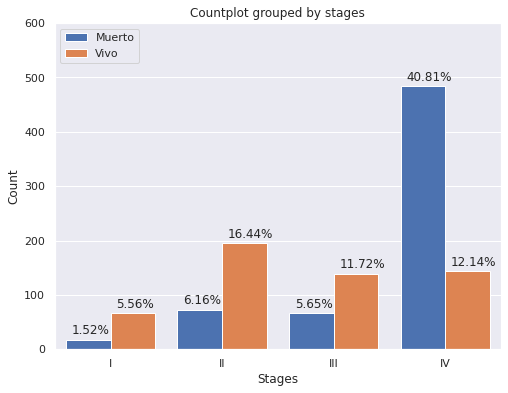
plt.figure(figsize = (8,6))
g=sns.countplot(x='E',
data=dp2.sort_values("E",ascending=True),
hue="Estado_Vital",hue_order=["Muerto","Vivo"],
saturation=1)
g.set(ylim=(0, 600))
g.set(title='Countplot grouped by stages')
g.set(xlabel='Stages', ylabel='Count');
plt.legend(loc='upper left')
for p in g.patches:
g.annotate('{:.2f}%'.format(p.get_height()*100/dp2['E'].count()), (p.get_x()+.05, p.get_height()+10))
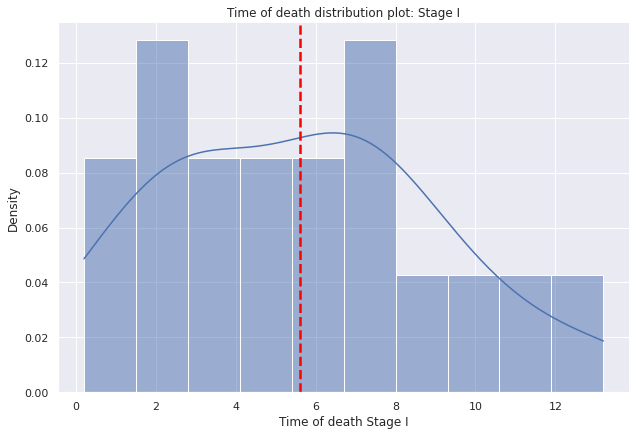
g=sns.displot(data=dp2[dp2["E"]=="I"],
x="tiempo_defuncion",
bins=10,kde=True,
kde_kws={'bw_adjust':0.9,'bw_method':'scott'},
stat='density', height=6,aspect=1.5).set(ylabel=None)
g.set(title='Time of death distribution plot: Stage I')
g.set(xlabel='Time of death Stage I', ylabel='Density')
plt.axvline(dp2[dp2["E"]=="I"]["tiempo_defuncion"].mean(),c="red", ls='--',lw=2.5);
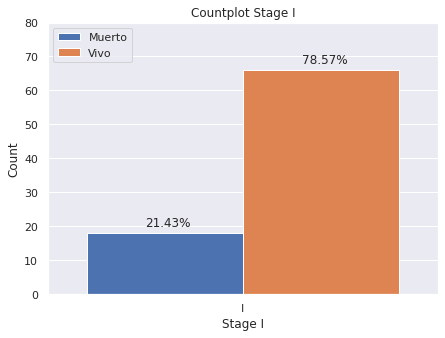
plt.figure(figsize = (7,5))
g=sns.countplot(x='E',
data=dp2[dp2["E"]=="I"],
hue="Estado_Vital",hue_order=["Muerto","Vivo"],
saturation=1)
g.set(ylim=(0, 80))
plt.legend(loc='upper left')
g.set(title='Countplot Stage I')
g.set(xlabel='Stage I', ylabel='Count')
for p in g.patches:
g.annotate('{:.2f}%'.format(p.get_height()*100/dp2[dp2["E"]=="I"]["index"].count()), (p.get_x()+.15, p.get_height()+2))
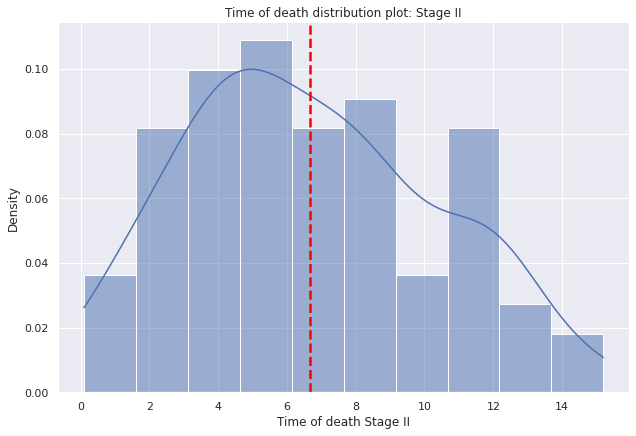
g=sns.displot(data=dp2[dp2["E"]=="II"],
x="tiempo_defuncion",
bins=10,kde=True,
kde_kws={'bw_adjust':0.9,'bw_method':'scott'},
stat='density', height=6,aspect=1.5).set(ylabel=None)
g.set(title='Time of death distribution plot: Stage II')
g.set(xlabel='Time of death Stage II', ylabel='Density')
plt.axvline(dp2[dp2["E"]=="II"]["tiempo_defuncion"].mean(),c="red", ls='--',lw=2.5);
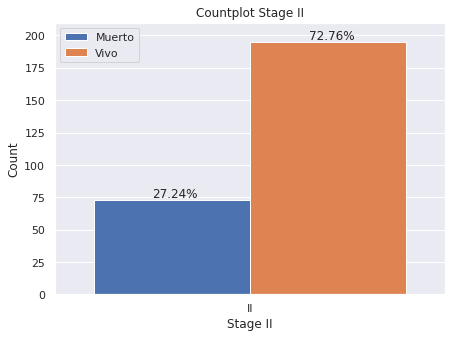
plt.figure(figsize = (7,5))
g=sns.countplot(x='E',
data=dp2[dp2["E"]=="II"],
hue="Estado_Vital",hue_order=["Muerto","Vivo"],
saturation=1)
g.set(ylim=(0, 210))
plt.legend(loc='upper left')
g.set(title='Countplot Stage II')
g.set(xlabel='Stage II', ylabel='Count')
for p in g.patches:
g.annotate('{:.2f}%'.format(p.get_height()*100/dp2[dp2["E"]=="II"]["index"].count()), (p.get_x()+.15, p.get_height()+2))
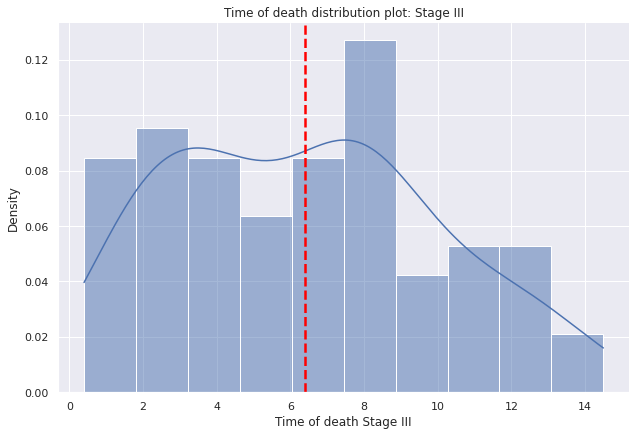
g=sns.displot(data=dp2[dp2["E"]=="III"],
x="tiempo_defuncion",
bins=10,kde=True,
kde_kws={'bw_adjust':0.9,'bw_method':'scott'},
stat='density', height=6,aspect=1.5).set(ylabel=None)
g.set(title='Time of death distribution plot: Stage III')
g.set(xlabel='Time of death Stage III', ylabel='Density')
plt.axvline(dp2[dp2["E"]=="III"]["tiempo_defuncion"].mean(),c="red", ls='--',lw=2.5);
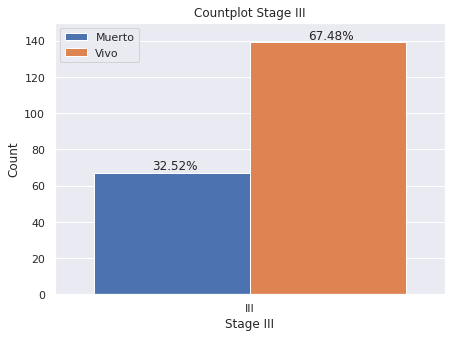
plt.figure(figsize = (7,5))
g=sns.countplot(x='E',
data=dp2[dp2["E"]=="III"],
hue="Estado_Vital",hue_order=["Muerto","Vivo"],
saturation=1)
g.set(ylim=(0, 150))
plt.legend(loc='upper left')
g.set(title='Countplot Stage III')
g.set(xlabel='Stage III', ylabel='Count')
for p in g.patches:
g.annotate('{:.2f}%'.format(p.get_height()*100/dp2[dp2["E"]=="III"]["index"].count()), (p.get_x()+.15, p.get_height()+2))
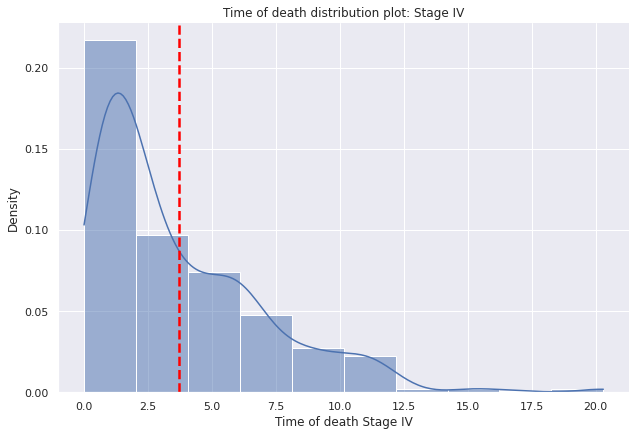
g=sns.displot(data=dp2[dp2["E"]=="IV"],
x="tiempo_defuncion",
bins=10,kde=True,
kde_kws={'bw_adjust':0.9,'bw_method':'scott'},
stat='density', height=6,aspect=1.5).set(ylabel=None)
g.set(title='Time of death distribution plot: Stage IV')
g.set(xlabel='Time of death Stage IV', ylabel='Density')
plt.axvline(dp2[dp2["E"]=="IV"]["tiempo_defuncion"].mean(),c="red", ls='--',lw=2.5);
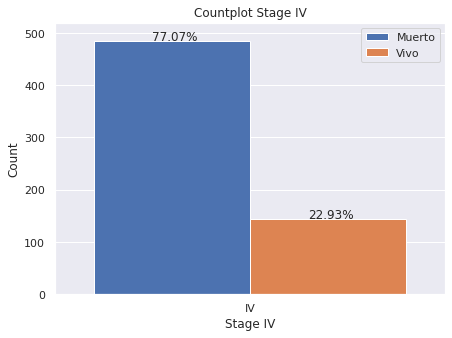
plt.figure(figsize = (7,5))
g=sns.countplot(x='E',
data=dp2[dp2["E"]=="IV"],
hue="Estado_Vital",hue_order=["Muerto","Vivo"],
saturation=1)
g.set(ylim=(0, 520))
plt.legend(loc='upper right')
g.set(title='Countplot Stage IV')
g.set(xlabel='Stage IV', ylabel='Count')
for p in g.patches:
g.annotate('{:.2f}%'.format(p.get_height()*100/dp2[dp2["E"]=="IV"]["index"].count()), (p.get_x()+.15, p.get_height()+2))
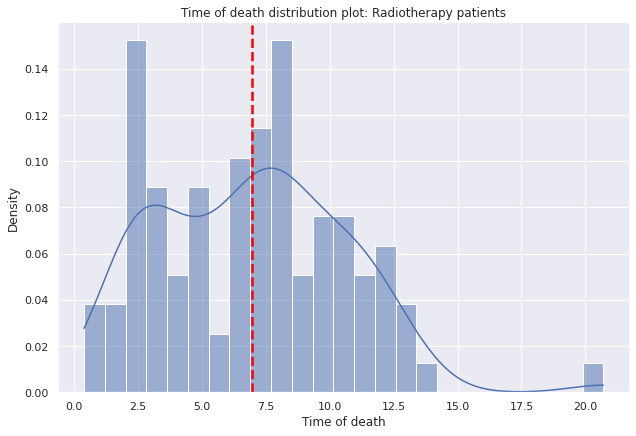
Radiotherapy
g=sns.displot(data=dp2[(dp2["Radioterapia"]==1)],
x="tiempo_defuncion",
bins=25,kde=True,
kde_kws={'bw_adjust':0.9,'bw_method':'scott'},
stat='density', height=6,aspect=1.5).set(ylabel=None)
g.set(title='Time of death distribution plot: Radiotherapy patients')
g.set(xlabel='Time of death', ylabel='Density')
plt.axvline(dp2[dp2["Radioterapia"]==1]["tiempo_defuncion"].mean(),c="red", ls='--',lw=2.5);
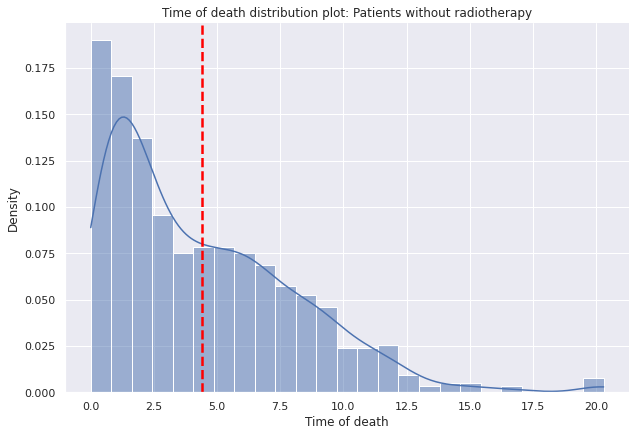
g=sns.displot(data=dp2[dp2["Radioterapia"]==0],x="tiempo_defuncion",
bins=25,kde=True,
kde_kws={'bw_adjust':0.9,'bw_method':'scott'},
stat='density', height=6,aspect=1.5).set(ylabel=None)
g.set(title='Time of death distribution plot: Patients without radiotherapy')
g.set(xlabel='Time of death', ylabel='Density')
plt.axvline(dp2[dp2["Radioterapia"]==0]["tiempo_defuncion"].mean(),c="red", ls='--',lw=2.5);
Survival
from lifelines import KaplanMeierFitter
from lifelines.utils import survival_events_from_table
from lifelines.utils import survival_table_from_events
df_survival=dp2.assign(R=dp2["Radioterapia"],
C=lambda dataset:dataset["Causa_Defuncion"].apply(lambda row:1 if row=="Otros" or row!=np.nan else 0),
S=lambda dataset:dataset["Estado_Vital"].apply(lambda row:1 if row=="Muerto"
else 0 if row=="Vivo"
else np.nan),
tiempo_vivo=dp2["tiempo_vivo"].replace(np.nan,""),
tiempo_defuncion=dp2["tiempo_defuncion"].replace(np.nan,""))
df_survival=df_survival.assign(T=lambda dataset:((dataset["tiempo_defuncion"].astype("str"))+dataset["tiempo_vivo"].astype("str")).astype("float"))
df_survival=df_survival.assign(T=round(df_survival["T"],0))
df_survival=df_survival[["T","S","C","E","R"]].dropna(subset=["E"])
df_survival.set_index("T",inplace=True,drop=False)
time, event, weight = survival_events_from_table(df_survival,
observed_deaths_col="S",
censored_col="C")
table=survival_table_from_events(df_survival["T"],
df_survival["S"])
print(table.head())
removed observed censored entrance at_risk
event_at
0.0 57 57 0 1186 1186
1.0 101 101 0 0 1129
2.0 109 109 0 0 1028
3.0 57 57 0 0 919
4.0 58 58 0 0 862
kmf=KaplanMeierFitter()
plt.figure(figsize = (8,6))
timelines=range(0,25,5)
kmf.fit(time,event,label="Survival Curve", timeline=timelines)
fig=kmf.plot_survival_function(show_censors=False)
fig.set(ylim=(0, 1.1),xlim=(0, 23.0))
fig.set_title('Survival Curve of all patients')
i=0
for item in kmf.survival_function_["Survival Curve"]:
if item>.5:
fig.annotate(str(round(item,2)),xy=(i+2,round(item,2)+.05))
else:
fig.annotate(str(round(item,2)),xy=(i+0.8,round(item,2)))
i+=5
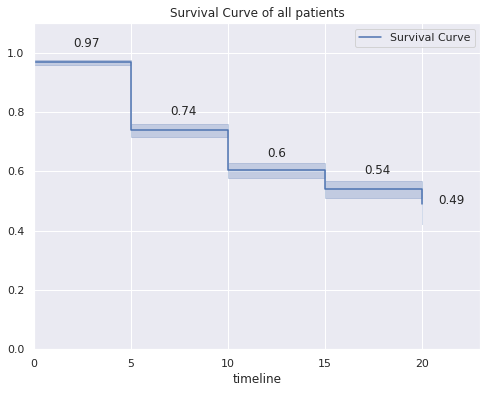
kmf.survival_function_
| Survival Curve | |
|---|---|
| timeline | |
| 0.0 | 0.968818 |
| 5.0 | 0.739353 |
| 10.0 | 0.603386 |
| 15.0 | 0.539839 |
| 20.0 | 0.490894 |
plt.figure(figsize = (8,6))
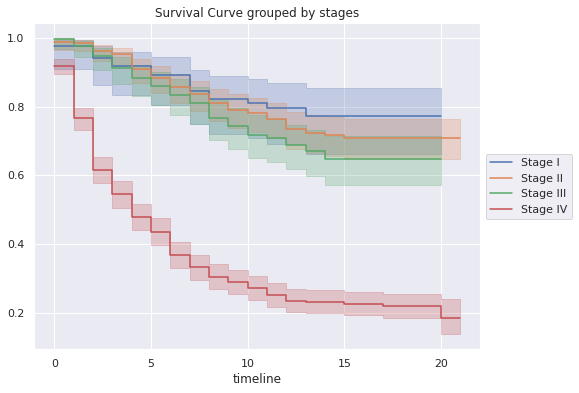
kmf.fit(df_survival[df_survival["E"]=="I"]["T"],
df_survival[df_survival["E"]=="I"]["S"],
label="Stage I")
df_s1=kmf.survival_function_
ax=kmf.plot()
kmf.fit(df_survival[df_survival["E"]=="II"]["T"],
df_survival[df_survival["E"]=="II"]["S"],
label="Stage II")
df_s2=kmf.survival_function_
ax=kmf.plot(ax=ax)
kmf.fit(df_survival[df_survival["E"]=="III"]["T"],
df_survival[df_survival["E"]=="III"]["S"],
label="Stage III")
df_s3=kmf.survival_function_
ax=kmf.plot(ax=ax)
kmf.fit(df_survival[df_survival["E"]=="IV"]["T"],
df_survival[df_survival["E"]=="IV"]["S"],
label="Stage IV")
df_s4=kmf.survival_function_
ax=kmf.plot(ax=ax)
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5));
ax.set_title('Survival Curve grouped by stages')
Text(0.5, 1.0, 'Survival Curve grouped by stages')
df_stages=pd.merge(df_s1,df_s2,on='timeline')
df_stages=pd.merge(df_stages,df_s3,on='timeline')
df_stages=pd.merge(df_stages,df_s4,on='timeline')
df_stages
| Stage I | Stage II | Stage III | Stage IV | |
|---|---|---|---|---|
| timeline | ||||
| 0.0 | 0.976190 | 0.988806 | 0.995146 | 0.918790 |
| 2.0 | 0.940476 | 0.962687 | 0.946602 | 0.616242 |
| 3.0 | 0.916667 | 0.951493 | 0.912621 | 0.544586 |
| 5.0 | 0.892857 | 0.884328 | 0.859223 | 0.436306 |
| 7.0 | 0.845238 | 0.835821 | 0.810680 | 0.332803 |
| 8.0 | 0.821429 | 0.809701 | 0.766990 | 0.305732 |
| 10.0 | 0.809524 | 0.779851 | 0.718447 | 0.272293 |
| 11.0 | 0.796032 | 0.763519 | 0.707394 | 0.250969 |
| 12.0 | 0.796032 | 0.735240 | 0.686988 | 0.235160 |
| 13.0 | 0.773288 | 0.723476 | 0.669596 | 0.232735 |
| 14.0 | 0.773288 | 0.716586 | 0.647996 | 0.232735 |
| 15.0 | 0.773288 | 0.708712 | 0.647996 | 0.225989 |
| 16.0 | 0.773288 | 0.708712 | 0.647996 | 0.225989 |
| 17.0 | 0.773288 | 0.708712 | 0.647996 | 0.220195 |
| 18.0 | 0.773288 | 0.708712 | 0.647996 | 0.220195 |
| 20.0 | 0.773288 | 0.708712 | 0.647996 | 0.186319 |
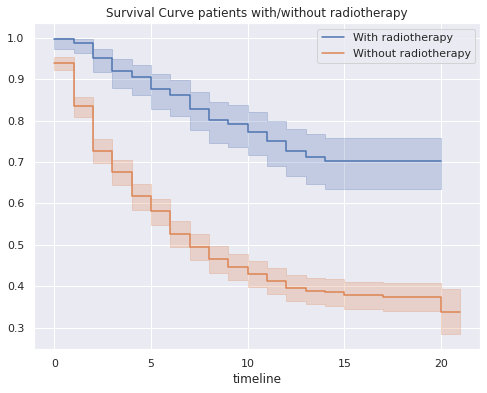
plt.figure(figsize = (8,6))
kmf.fit(df_survival[df_survival["R"]==1]["T"],df_survival[df_survival["R"]==1]["S"],label="With radiotherapy")
df_R1=kmf.survival_function_
ax=kmf.plot()
kmf.fit(df_survival[df_survival["R"]==0]["T"],df_survival[df_survival["R"]==0]["S"],label="Without radiotherapy")
df_R0=kmf.survival_function_
ax=kmf.plot(ax=ax)
ax.set_title('Survival Curve patients with/without radiotherapy');
df_R=pd.merge(df_R1,df_R0,on='timeline')
df_R
| With radiotherapy | Without radiotherapy | |
|---|---|---|
| timeline | ||
| 0.0 | 0.996016 | 0.940107 |
| 1.0 | 0.988048 | 0.834225 |
| 2.0 | 0.952191 | 0.727273 |
| 3.0 | 0.920319 | 0.674866 |
| 4.0 | 0.904382 | 0.617112 |
| 5.0 | 0.876494 | 0.580749 |
| 6.0 | 0.860558 | 0.527273 |
| 7.0 | 0.828685 | 0.495187 |
| 8.0 | 0.800797 | 0.465241 |
| 9.0 | 0.792829 | 0.445989 |
| 10.0 | 0.772908 | 0.429947 |
| 11.0 | 0.749767 | 0.412975 |
| 12.0 | 0.727218 | 0.395601 |
| 13.0 | 0.711579 | 0.389089 |
| 14.0 | 0.701963 | 0.385255 |
| 15.0 | 0.701963 | 0.378293 |
| 16.0 | 0.701963 | 0.378293 |
| 17.0 | 0.701963 | 0.374225 |
| 18.0 | 0.701963 | 0.374225 |
| 19.0 | 0.701963 | 0.374225 |
| 20.0 | 0.701963 | 0.338585 |
Recurrence
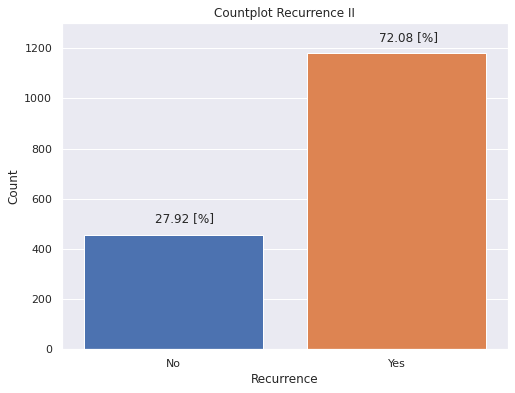
plt.figure(figsize = (8,6))
g=sns.countplot(x='Recurrecia',
data=dp2,
saturation=1)
g.set(ylim=(0, 1300))
labels = (["No","Yes"])
g.set_xticklabels(labels)
g.set(xlabel='Recurrence', ylabel='Count')
g.set(title='Countplot Recurrence II')
for p in g.patches:
g.annotate('{:.2f} [%]'.format(p.get_height()*100/dp2['Edad_diag'].count()), (p.get_x()+0.32, p.get_height()+50))
df_recurr=dp2.assign(C=lambda dataset:dataset["Aband_Tto"],
S=lambda dataset:dataset["Recurrecia"])
df_recurr=df_recurr.assign(T=round(df_recurr["tiempo_recurr"],0))
df_recurr=df_recurr[["T","S","C","E"]].dropna(subset=["E"])
df_recurr.replace(np.nan,0,inplace=True)
df_recurr.set_index("T",inplace=True,drop=False)
time, event, weight = survival_events_from_table(df_recurr,
observed_deaths_col="S",
censored_col="C")
table=survival_table_from_events(df_recurr["T"],
df_recurr["S"])
print(table.head())
removed observed censored entrance at_risk
event_at
0.0 380 50 330 1186 1186
1.0 95 95 0 0 806
2.0 90 90 0 0 711
3.0 44 44 0 0 621
4.0 47 47 0 0 577
kmf2=KaplanMeierFitter()
plt.figure(figsize = (8,6))
timelines=range(0,25,5)
kmf2.fit(time,event,label="Recurrence Curve", timeline=timelines)
fig=kmf2.plot_survival_function(show_censors=False)
fig.set(ylim=(0, 1.1),xlim=(0, 23.0))
fig.set_title('Recurrence Curve of all patients')
i=0
for item in kmf2.survival_function_["Recurrence Curve"]:
if item>.03:
fig.annotate(str(round(item,2)),xy=(i+2,round(item,2)+.05))
else:
fig.annotate(str(round(item,2)),xy=(i+0.8,round(item,2)))
i+=5
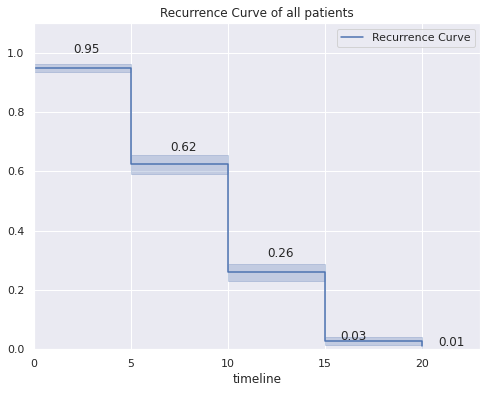
kmf2.survival_function_
| Recurrence Curve | |
|---|---|
| timeline | |
| 0.0 | 0.949495 |
| 5.0 | 0.623724 |
| 10.0 | 0.259126 |
| 15.0 | 0.026448 |
| 20.0 | 0.011652 |
plt.figure(figsize = (8,6))
kmf2.fit(df_recurr[df_recurr["E"]=="I"]["T"],df_recurr[df_recurr["E"]=="I"]["S"],label="Stage I")
df2_s1=kmf2.survival_function_
ax=kmf2.plot()
kmf2.fit(df_recurr[df_recurr["E"]=="II"]["T"],df_recurr[df_recurr["E"]=="II"]["S"],label="Stage II")
df2_s2=kmf2.survival_function_
ax=kmf2.plot(ax=ax)
kmf2.fit(df_recurr[df_recurr["E"]=="III"]["T"],df_recurr[df_recurr["E"]=="III"]["S"],label="Stage III")
df2_s3=kmf2.survival_function_
ax=kmf2.plot(ax=ax)
kmf2.fit(df_recurr[df_recurr["E"]=="IV"]["T"],df_recurr[df_recurr["E"]=="IV"]["S"],label="Stage IV")
df2_s4=kmf2.survival_function_
ax=kmf2.plot(ax=ax)
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5));
ax.set_title('Recurrence Curve grouped by stages')
Text(0.5, 1.0, 'Recurrence Curve grouped by stages')
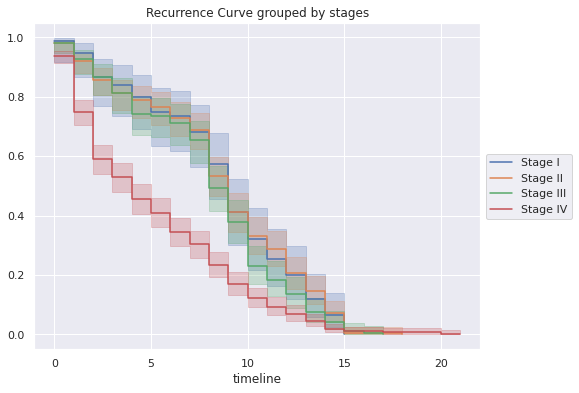
df_stages_recurr=pd.merge(df2_s1,df2_s2,on='timeline')
df_stages_recurr=pd.merge(df_stages_recurr,df2_s3,on='timeline')
df_stages_recurr=pd.merge(df_stages_recurr,df2_s4,on='timeline')
df_stages_recurr
| Stage I | Stage II | Stage III | Stage IV | |
|---|---|---|---|---|
| timeline | ||||
| 0.0 | 0.988095 | 0.981343 | 0.980583 | 0.936306 |
| 1.0 | 0.948037 | 0.922006 | 0.927096 | 0.750109 |
| 2.0 | 0.867921 | 0.858105 | 0.867667 | 0.590511 |
| 3.0 | 0.841216 | 0.812461 | 0.814181 | 0.529332 |
| 4.0 | 0.801158 | 0.789639 | 0.742866 | 0.457513 |
| 5.0 | 0.747748 | 0.766817 | 0.736923 | 0.409634 |
| 6.0 | 0.734395 | 0.730302 | 0.713151 | 0.345795 |
| 7.0 | 0.680985 | 0.689222 | 0.653722 | 0.303235 |
| 8.0 | 0.574163 | 0.534033 | 0.493263 | 0.234076 |
| 9.0 | 0.413932 | 0.410795 | 0.380347 | 0.170237 |
| 10.0 | 0.320463 | 0.333200 | 0.231774 | 0.122358 |
| 11.0 | 0.253700 | 0.287556 | 0.184231 | 0.093099 |
| 12.0 | 0.200290 | 0.205397 | 0.136687 | 0.069159 |
| 13.0 | 0.120174 | 0.146060 | 0.077258 | 0.045219 |
| 14.0 | 0.066763 | 0.073030 | 0.041600 | 0.018620 |
| 15.0 | 0.000000 | 0.004564 | 0.011886 | 0.010640 |
Conclusions
- The average age at diagnosis of prostate cancer in the sample was around 70 years.
- Patients with stage IV disease have a median age of 72 years, while patients with stages I, II and III have a median age greater than 65 and less than 70 years.
- The mean age at death of the patients in the sample who have died is around 5 years, which means that the majority, at least on average, live 5 years.
- The average time of death of patients is around 5 years.
- The median time of patients alive in 2020 is around 14-15 years.
- More than 50% of the patients in the sample have been diagnosed with stage IV.
- De la muestra de pacientes agrupados por estadío clínico, se pude observar:
- Estadio I: 78.57% de los pacientes esta vivo hasta el momento de la generación de la base de datos en 2020
- Estadio II: 72.76% de los pacientes esta vivo hasta el momento de la generación de la base de datos en 2020
- Estadio III: 67.48% de los pacientes esta vivo hasta el momento de la generación de la base de datos en 2020
- Estadio IV: 22.93% de los pacientes esta vivo hasta el momento de la generación de la base de datos en 2020
- Patients who have received radiotherapy, at least on average, have a longer average lifespan than those who have not received radiotherapy.
- About the overall survival of the patients in the sample:
- The probability of survival of a patient in the first 5 years after being diagnosed is 0.97.
- The probability of survival of a patient between 5 and 10 years after diagnosis is 0.74
- The probability of survival of a patient between 10 and 15 years after diagnosis is 0.6
- The probability of survival of a patient between 15 and 20 years after diagnosis is 0.54
- The probability of survival of a patient more than 20 years after diagnosis is 0.49
- About the survival grouped by stage of the patients in the sample:
- The 5-year survival probability of a patient presenting stage I is 0.89, while a patient presenting stage IV is 0.44.
- The 10-year survival probability of a patient presenting with stage I is 0.81, while a patient presenting with stage IV is 0.27.
- The 15-year survival probability for a patient presenting with stage I is 0.77, while a patient presenting with stage IV is 0.23.
- The 20-year survival probability for a patient presenting with stage I is 0.77, while a patient presenting with stage IV is 0.19.
- The probability of survival of a patient with stage I, II and II is much higher than that of a patient with stage IV.
- As the patient is diagnosed at an advanced stage of the disease, the probability of survival decreases dramatically.
- On the survival of the sample grouped by those patients who received radiotherapy:
- The 5-year survival probability of a patient who received radiotherapy is 0.88, while that of a patient who did not receive is 0.58.
- The 10-year survival probability of a patient who received radiotherapy is 0.77, while that of a patient who did not receive is 0.43.
- The 15-year survival probability of a patient who received radiotherapy is 0.70, while that of a patient who did not receive is 0.38.
- The 20-year survival probability of a patient who received radiotherapy is 0.70, while that of a patient who did not receive is 0.34.
- There is a large difference in the survival probability of patients who received radiotherapy, which varies greatly depending on those who did not receive this treatment.
- About the recurrence of patients:
- 78.08% of patients present a recurrence of cancer during the study time.
- The probability that the disease does not recur in the first 5 years is 0.95
- The probability that the disease will not recur in the first 10 years is 0.62%.
- The probability that the disease does not recur in the first 15 years is 0.26
- The probability that the disease will not recur within the first 20 years is 0.03
- The probability that the disease will not recur in the first 20 years is 0.01.
- The probability of non-recurrence in patients with stage IV is much lower than in those with stages I, II and III, but the probability of these stages decreasing together after 10 years, with stages I and II being very similar.